Lecture 7Effects of ligands on redox potential |
Redox proteins usually function through oxidation reduction reactions at a local site. Most commonly, the site is a bound coenzyme, or a prosthetic group, but occasionally an amino acid side chain is used; several examples of these different options are included on the redox components page. There are relatively few common prosthetic groups, so many redox proteins have the same redox center, but very different redox properties. In order to understand how the redox potential of a center can be modified by its binding site, it is useful to consider the simple general case of how a ligand changes the redox potential of a half-cell reaction.
When a ligand binds to a chemical species, the activity (concentration) of that species is less because it is removed from the reaction mix. When a ligand binds to a redox component, the apparent redox potential will change if the ligand binds differentially to the oxidized and reduced components of the half-cell reaction. We can represent this formally through the following equations:
Eo'
[A-][L]
KR = -------
[A-L]
[A][L]
KR = -------
[AL]
E1o'
E2o'
E3o'
Note, each redox half-cell (equations (i), (iv), (v), and (vi)) has a different midpoint potential. Note also that the equilibrium constants are dissociation constants.
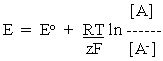 -----(1)
-----(1)
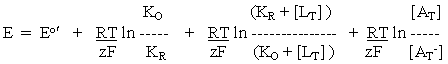 -----(2)
-----(2)
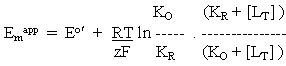 -----(3)
-----(3)
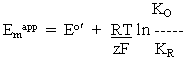 -----(4)
-----(4)Note also that these relationships are reversed if the equilibrium constants used are association constants, rather than the dissociation constants used here, as appropriate for the reactions shown in eq. (ii) and (iii) above.